![[Prev]](p_prev.gif)
![[Next]](p_next.gif)
![[Prev]](p_prev.gif)
![[Next]](p_next.gif)
# Program rules.tcl
proc Tcl_rule {} {
###################################
# Procedure : Tcl_rule
# Arguments :
# irule - rule number
# op - part of the used rule
# sub_op - number of the model function
# Returns :
# OK , BAD
#
####################################
global irule op sub_op rule_trace
global_name
global kp1 c_h2o c_h2
global name date sconc kin_mode conversion_limit reaction_time
global minimal_concentration
global phases_p_r phase_contacts phprop comp_edus in_phase phase
global attrib_1 center_1 rule_1
global attrib_2 center_2 rule_2
global attrib_3 center_3 rule_3
global attrib_4 center_4 rule_4
switch $irule {
# Regelkopf
GLOBAL {
switch $op {
# Einstellungen der Reaktoren, Phasen, Kinetik
INIT_RULES {
set c_h2o 1.0
set c_h2 1.0
set name nt_tria09.tcl"
putco name name
set date 27.06.1997
get temperature temp
print temperature $temp"
putco date date
# reactors and phases
set in_phase(0) 0
set comp_edus(0) 1
set phases_p_r {1,0}
set phase_contacts(0) 0
set phprop(0) $MONOMOLEC"
putco phases_per_reactor phases_p_r
putco phase_contacts phase_contacts
putco phase_property phprop
putco use_all_educts_together comp_edus
putco input_phase_for_reactors in_phase
set phase 1
putco output_phase phase
# kinetic
set sconc(0) 0.10
set conversion_limit 0.0
set reaction_time 10368.e3
set minimal_concentration 1.e-10
set kin_mode(0) gear
putco kinetic_model kin_mode
putco conversion_limit conversion_limit
putco reaction_time reaction_time
putco minimal_concentration minimal_concentration
put start_conc sconc
# give rule variables to kernel
set kp1 0.0
put reactivity kp1
print test $test trace $trace"
print INIT_RULES called"
return OK
}
# Vorbehandlung der Ausgangsmaterialien
PREP_ROOT {
print PREP_ROOT called .
return OK
}
# Vorbehandlung der Edukte
PREP_EDUCT {
print PREP_EDUCT called ."
return OK
}
# Verteilungsfunktion
DISTRIBFUNC {
return OK
}
# Abschlußfunktion
FINISH {
return OK
}
default {
return BAD
}
}
}
# Allgemein Hydrolyseregel
RULE_1 {
switch $op {
# Definition des Namen, der Attribute und der RSS
RULE_INFO {
set rule_1 Allg. Hydrolyse"
putco rule_name rule_1
set attrib_1(0) NULL
putco attributes attrib_1
set center_1 {1,2,0,2,3,0,0}
putco center_connectivity center_1
set ff [net cpg init trained.knet test3-1+0.ctxeda]
if {!$ff} {
print Rule 1: error during the net initialization
}
return OK
}
# Überprüfung der Bedingungen für die RSS
CONSTR {
switch $sub_op {
GLOBAL {
return OK
}
# für das Eduktensemble
0 {
if {$trace>2} {
print Rule 1: Checking total ensemble
print trace=$trace"
}
set blt [prop E_N_ATOMS na]
if {!$blt} {
print error getting property E_N_ATOMS"
}
set blt [prop E_LAST_ATOM il]
if {!$blt} {
print error getting property E_LAST_ATOM
}
set sar 0
for {set i 1} {$i<=$il} {incr i} {
set blt [prop A_AROMATIC $i aromat]
if {$blt} {
if {$aromat} {incr sar}
}
}
set ar $sar
if {$ar!=6&&$na>7} {return BAD}
return OK
}
# für das erste Atom in der RSS
1 {
if {$trace>2} {
print Checking atom 1 rule 1
}
set atom [center 1]
set blt [prop A_ELEMENT $atom elem]
if {!$blt} {
print error getting property A_ELEMENT $atom"
}
if {$elem!=6&&$elem!=8&&$elem!=7&&$elem!=9&&$elem!=17&&$elem!=35&&$elem!=35&&$elem!=16} {return BAD}
return OK
}
# für das zweite Atom in der RSS
2 {
if {$trace>2} {
print Checking atom 2 rule 1
}
set atom [center 2]
set blt [prop A_ELEMENT $atom elem]
if {!$blt} {
print error getting property A_ELEMENT $atom"
}
if {$elem!=6} {return BAD}
return OK
}
3 {
if {$trace>2} {
print Checking atom 3 rule 1
}
set atom1 [center 1]
set atom2 [center 2]
set atom3 [center 3]
set blt [prop A_ELEMENT $atom1 elem1]
if {!$blt} {
print error getting property A_ELEMENT $atom1"
}
set blt [prop A_ELEMENT $atom2 elem2]
if {!$blt} {
print error getting property A_ELEMENT $atom2"
}
set blt [prop A_ELEMENT $atom3 elem3]
if {!$blt} {
print error getting property A_ELEMENT $atom1"
}
if {$elem3!=6&&$elem3!=8&&$elem3!=1&&$elem3!=16&&$elem3!=7} {return BAD}
set blt [prop A_ENSIG $atom2 ensig2]
if {!$blt} {
print error getting property A_ENSIG $atom2"
}
set blt [prop A_NEIGHBORS $atom1 ng]
if {!$blt} {
print error getting property A_NEIGHBORS $atom1"
}
set ll [array size ng]
set nhneigh1 0
for {set i 0} {$i<$ll} {incr i} {
set blt [prop A_ELEMENT $ng($i) j]
if {!$blt} {
print error getting property A_ELEMENT $ng($i)"
}
if {$j!=1} {incr nhneigh1}
}
set blt [prop B_BOORD $atom1 $atom2 bor1]
if {!$blt} {
print error getting property B_BOORD $atom1 $atom2"
}
set blt [prop B_BOORD $atom2 $atom3 bor2]
if {!$blt} {
print error getting property B_BOORD $atom2 $atom3"
}
set blt [prop A_AROMATIC $atom1 ar1]
if {!$blt} {
print error getting property A_AROMATIC $atom1"
}
set blt [prop A_AROMATIC $atom2 ar2]
if {!$blt} {
print error getting property A_AROMATIC $atom2"
}
if {$trace>3} {
print Rule 1: e1=$elem1 e3=$elem3 ensig=$ensig2 bor1=$bor1 ar1=$ar1 ar2=$ar2 nhneigh1=$nhneigh1
}
if {$elem1==7&&$elem3==6&&$ar2!=0} {return BAD}
if {$elem1==8&&$bor1>1} {return BAD}
if {$bor1==1&&$elem1==7&&$elem3==8&&$ensig2<10.1} {return BAD}
if {$elem1==7&&$elem3==7&&$ar2!=0&&$nhneigh1>1} {return BAD}
if {$elem1==6&&$elem2==6} {return BAD}
if {$ar1!=0} {return BAD}
if {$elem1==8&&$nhneigh1==1} {return BAD}
if {$elem1==7&&$ar2==0&&$elem3!=8&&$elem3!=7} {return BAD}
return OK
}
default {
print illegal number of atoms"
return BAD
}
}
}
# Reaktionsfunktion mit Festlegung der reaktivität
FUNC {
# Eduktfunktion
if {$trace>2} {
print Rule 1: FUNC called
}
set atom1 [center 1]
set atom2 [center 2]
set atom3 [center 3]
set ff [net cpg set educt]
if {!$ff} {
print Rule 1: net cpg set educt failed."
}
# Durchführung der Reaktion
set h1 [new_atom 1 elsys]
set h2 [new_atom 1 elsys]
set o [new_atom 8 elsys]
change_bond_order $atom2 $atom1 -1
change_bond_order $h1 $o 1
change_bond_order $h2 $atom1 1
change_bond_order $o $atom2 1
# Produktfunktion
set fff [net cpg set product]
if {!$fff} {
print net cpg set product failed."
}
if {$ff&&$fff} {
set f1 [net cpg get R1]
if {$f1} {
set ar [array exist R1]
if {$ar} {
set dim [array size R1]
for {set i 0} {$i<$dim} {incr i} {
print R1[$i]=$R1[$i]"
}
} else {
print R1=$R1"
}
} else {
print error getting the net result"
}
} else {
print error setting the net values"
}
# Überprüfung, ob wirklich alles paßt
set blt [prop A_ELEMENT $atom1 elem]
if {!$blt} {
print error getting property A_ELEMENT $atom1"
}
set blt [prop A_ELEMENT $atom3 elem3]
if {!$blt} {
print error getting property A_ELEMENT $atom3"
}
set blt [prop B_BOORD $atom1 $atom2 bor1]
if {!$blt} {
print error getting property B_BOORD $atom1 $atom2"
}
set blt [prop A_AROMATIC $atom2 ar2]
if {!$blt} {
print error getting property A_AROMATIC $atom2"
}
if {$elem==7&&$ar2==0&&$bor1==0&&$elem3!=8&&$elem3!=7} {return BAD}
# konjugiere pi-Systeme
set blt [prop A_ELECSYSS $atom2 esys2]
if {!$blt} {
print error getting property A_ELECSYSS $atom2"
}
set blt [prop A_ELECSYSS $o esyso]
if {!$blt} {
print error getting property A_ELECSYSS $o"
}
set picount1 0
set dim [array size esys2]
for {set i 0} {$i<$dim} {incr i} {
set blt [prop EL_IS_SIGMA $esys2($i) is_sigma]
if {!$blt} {
print error getting property EL_IS_SIGMA $esys2($i)"
}
if {!$is_sigma} {
set pis2 $esys2($i)
incr picount1
}
}
set picount2 0
set dim [array size esyso]
for {set i 0} {$i<$dim} {incr i} {
set blt [prop EL_IS_SIGMA $esyso($i) is_sigma]
if {!$blt} {
print error getting property EL_IS_SIGMA $esyso($i)"
}
if {!$is_sigma} {
set piso $esyso($i)
incr picount2
}
}
if {$picount1&&$picount2} {
set blt [combine_elsys $pis2 $piso es_pi]
if {!$blt} {
print combine_elsys $pis2 $piso"
}
print combine c-o"
}
# the end
set kp1 [expr 5.e-9*$c_h2o]
set symmetry_factor [expr $symmetry_factor*2]
if {$trace>2} {
print symmetry_factor=$symmetry_factor"
}
return OK
}
default {
return BAD
}
}
}
# Reaktionstyp der reduktiven Dealkylierung
RULE_2 {
switch $op {
# Name, Attribute, RSS
RULE_INFO {
set rule_2 Reduktive Dealkylierung"
putco rule_name rule_2
set attrib_2(0) NULL
putco attributes attrib_2
set center_2 {1,2,0,0}
putco center_connectivity center_2
return OK
}
# Bedingungen der RSS
CONSTR {
switch $sub_op {
GLOBAL {
return OK
}
0 {
if {$trace>2} {
print Checking total ensembles rule 2
}
set blt [prop E_LAST_ATOM il]
if {!$blt} {
print error getting property E_LAST_ATOM
}
set sar 0
for {set i 1} {$i<=$il} {incr i} {
set blt [prop A_AROMATIC $i aromat]
if {$blt} {
if {$aromat} {incr sar}
}
}
set ar $sar
if {$ar!=6} {return BAD}
return OK
}
1 {
if {$trace>2} {
print Checking atom 1 rule 2"
}
set atom [center 1]
set blt [prop A_ELEMENT $atom elem]
if {!$blt} {
print error getting property A_ELEMENT $atom"
}
if {$elem!=7} {return BAD}
set blt [prop A_AROMATIC $atom ar]
if {!$blt} {
print error getting property A_AROMATIC $atom"
}
if {$ar!=0} {return BAD}
return OK
}
2 {
if {$trace>2} {
print Checking atom 2 rule 2"
}
set atom [center 2]
set blt [prop A_ELEMENT $atom elem]
if {!$blt} {
print error getting property A_ELEMENT $atom"
}
if {$elem!=6} {return BAD}
set blt [prop A_AROMATIC $atom ar]
if {!$blt} {
print error getting property A_AROMATIC $atom"
}
if {$ar!=0} {return BAD}
return OK
}
default {
print illegal number of atoms"
}
}
}
# Reaktionsfunktion
FUNC {
# Eduktfunktion
if {$trace>2} {
print FUNC called Rule 2"
}
# Durchführung der Reaktion
set atom1 [center 1]
set atom2 [center 2]
set h1 [new_atom 1 elsys]
set h2 [new_atom 1 elsys]
change_bond_order $atom2 $atom1 -1
change_bond_order $h1 $atom1 1
change_bond_order $h2 $atom2 1
# Produktfunktion
set blt [prop A_NEIGHBORS $atom2 ng]
if {!$blt} {
print error getting property A_AROMATIC $atom"
}
set ll [array size ng]
set nhneigh 0
for {set i 0} {$i<$ll} {incr i} {
set blt [prop A_ELEMENT $ng($i) j]
if {!$blt} {
print error getting property A_AROMATIC $atom"
}
if {$j!=1} {incr nhneigh}
}
set kp1 [expr $c_h2*4.42e-8*3./(2.+$nhneigh)]
set symmetry_factor [expr $symmetry_factor*2]
if {$trace>2} {
print symmetry_factor=$symmetry_factor kp1=$kp1"
}
return OK
}
default {
return BAD
}
}
}
# kombinierte Hydrolyse/Debarboxylierung für die Abbauprodukte der Cyanursäure
RULE_4 {
switch $op {
# Name, Attribute, RSS
RULE_INFO {
set rule_4 Decarboxylierung"
putco rule_name rule_4
set attrib_4(0) NULL
putco attributes attrib_4
set center_4 {1,2,0,2,3,0,3,4,0,4,5,0,5,6,0,4,7,0,0}
putco center_connectivity center_4
return OK
}
# Bedingungen der RSS
CONSTR {
switch $sub_op {
GLOBAL {
return OK
}
0 {
return OK
}
1 {
if {$trace>2} {
print Checking atom 1 rule 4
}
set atom [center 1]
set blt [prop A_ELEMENT $atom elem]
if {!$blt} {
print error getting property A_ELEMENT $atom"
}
if {$elem!=7} {return BAD}
return OK
}
2 {
if {$trace>2} {
print Checking atom 2 rule 4
}
set atom [center 2]
set blt [prop A_ELEMENT $atom elem]
if {!$blt} {
print error getting property A_ELEMENT $atom"
}
if {$elem!=6} {return BAD}
return OK
}
3 {
if {$trace>2} {
print Checking atom 3 rule 4
}
set atom3 [center 3]
set atom2 [center 2]
set blt [prop A_ELEMENT $atom3 elem]
if {!$blt} {
print error getting property A_ELEMENT $atom3"
}
if {$elem!=7} {return BAD}
set blt [prop B_BOORD $atom2 $atom3 bor2]
if {!$blt} {
print error getting property B_BOORD $atom2 $atom3"
return BAD
}
if {$bor2<2} {return BAD}
return OK
}
4 {
if {$trace>2} {
print Checking atom 4 rule 4
}
set atom4 [center 4]
set blt [prop A_ELEMENT $atom4 elem]
if {!$blt} {
print error getting property A_ELEMENT $atom4"
}
if {$elem!=6} {return BAD}
return OK
}
5 {
if {$trace>2} {
print Checking atom 5 rule 4
}
set atom5 [center 5]
set blt [prop A_ELEMENT $atom5 elem]
if {!$blt} {
print error getting property A_ELEMENT $atom5"
}
if {$elem!=8} {return BAD}
return OK
}
6 {
if {$trace>2} {
print Checking atom 6 rule 4
}
set atom6 [center 6]
set blt [prop A_ELEMENT $atom6 elem]
if {!$blt} {
print error getting property A_ELEMENT $atom6"
}
if {$elem!=1} {return BAD}
return OK
}
7 {
if {$trace>2} {
print Checking atom 7 rule 4
}
set atom4 [center 4]
set atom7 [center 7]
set blt [prop A_ELEMENT $atom7 elem]
if {!$blt} {
print error getting property A_ELEMENT $atom7"
}
if {$elem!=8} {return BAD}
set blt [prop B_BOORD $atom4 $atom7 bor]
if {!$blt} {
print error getting property B_BOORD $atom4 $atom7"
return BAD
}
if {$bor<2} {return BAD}
return OK
}
default {
print illegal number of atoms"
return BAD
}
}
}
# Reaktionsfunktion
FUNC {
# Eduktfunktion
if {$trace>2} {
print FUNC called Rule 4"
}
set c1 [center 1]
set c2 [center 2]
set c3 [center 3]
set c4 [center 4]
set c5 [center 5]
set c6 [center 6]
set c7 [center 7]
# Durchführung der Reaktion
set h1 [new_atom 1 elsys]
set h2 [new_atom 1 elsys]
set o [new_atom 8 elsys]
change_bond_order $c5 $c6 -1
change_bond_order $c2 $c3 -2
change_bond_order $c3 $c4 -1
change_bond_order $c2 $o 2
change_bond_order $c3 $h1 1
change_bond_order $c3 $h2 1
change_bond_order $c3 $c6 1
change_bond_order $c4 $c5 1
# Produktfunktion
set kp1 [expr 1.e-5*$c_h2o]
if {$trace>2} {
print symmetry_factor=$symmetry_factor kp1=$kp1"
}
return OK
}
default {
return BAD
}
}
}
# Hydrolyse von Cyanursäure
RULE_3 {
switch $op {
# Name, Attribute, RSS
RULE_INFO {
set rule_3 Hydrolyse von Cyanursaeure"
putco rule_name rule_3
set attrib_3(0) NULL
putco attributes attrib_3
set center_3 {1,2,0,2,3,0,3,4,0,0}
putco center_connectivity center_3
return OK
}
# Bedingungen der RSS
CONSTR {
switch $sub_op {
GLOBAL {
return OK
}
0 {
if {$trace>2} {
print Checking total ensemble rule 3"
}
# funktion ens_aromat
set blt [prop E_LAST_ATOM il]
if {!$blt} {
print error getting property E_LAST_ATOM
}
set sar 0
for {set i 1} {$i<=$il} {incr i} {
set blt [prop A_AROMATIC $i aromat]
if {$blt} {
if {$aromat} {incr sar}
}
}
set ar $sar
if {$ar!=6} {return BAD}
return OK
}
1 {
if {$trace>2} {
print Checking atom 1 rule 3
}
set atom [center 1]
set blt [prop A_ELEMENT $atom elem]
if {!$blt} {
print error getting property A_ELEMENT $atom"
}
if {$elem!=7} {return BAD}
return OK
}
2 {
if {$trace>2} {
print Checking atom 2 rule 3
}
set atom2 [center 2]
set atom1 [center 1]
set blt [prop A_ELEMENT $atom2 elem]
if {!$blt} {
print error getting property A_ELEMENT $atom"
}
if {$elem!=6} {return BAD}
set blt [prop B_BOORD $atom1 $atom2 bor1]
if {!$blt} {
print error getting property B_BOORD $atom1 $atom2"
}
set blt [prop A_ENSIG $atom2 esig]
if {!$blt} {
print error getting property A_ENSIG $atom2"
}
set blt [prop A_POLARIZ $atom2 apol]
if {!$blt} {
print error getting property A_ELEMENT $atom2"
}
# if {$bor1<2} {return BAD}
if {$esig<11.44||$esig>11.46} {return BAD}
if {$apol<5.9||$apol>6.1} {return BAD}
return OK
}
3 {
if {$trace>2} {
print Checking atom 3 rule 3
}
set atom3 [center 3]
set blt [prop A_ELEMENT $atom3 elem]
if {!$blt} {
print error getting property A_ELEMENT $atom3"
}
if {$elem!=8} {return BAD}
return OK
}
4 {
if {$trace>2} {
print Checking atom 4 rule 3
}
set atom4 [center 4]
set blt [prop A_ELEMENT $atom4 elem]
if {!$blt} {
print error getting property A_ELEMENT $atom3"
}
if {$elem!=1} {return BAD}
return O
}
default {
print illegal number of atoms"
return BAD
}
}
}
# Reaktionsfunktion
FUNC {
# Eduktfunktion
if {$trace>2} {
print FUNC called rule 3
}
set atom1 [center 1]
set atom2 [center 2]
set atom3 [center 3]
set atom4 [center 4]
# Durchführung der Reaktion
set h1 [new_atom 1 elsys]
set h2 [new_atom 1 elsys]
set o [new_atom 8 elsys]
change_bond_order $h1 $o 1
change_bond_order $atom1 $atom2 -2
change_bond_order $atom3 $atom4 -1
change_bond_order $h2 $atom1 1
change_bond_order $atom1 $atom4 1
change_bond_order $o $atom2 1
change_bond_order $atom2 $atom3 1
# Produktfunktion
set kp1 [expr 1.e-6*$c_h2o]
if {$trace>2} {
print symmetry_factor=$symmetry_factor kp1=$kp1"
}
return OK
}
default {
return BAD
}
}
}
# end of the rules
default {
print illegal rule number"
return BAD
}
}
}
# Eine zweite Regelfuntkion
proc Test_rule {} {
global_name
print hier koennten weitere Regeln stehen"
return BAD
}
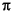 -System konjugiert wird, ist Absicht, da so auf den Mechanismus zur Verhinderung der doppelten Wasserstoffumlagerung auf eine funktionelle Gruppe verzichtet werden konnte, da die
-System konjugiert wird, ist Absicht, da so auf den Mechanismus zur Verhinderung der doppelten Wasserstoffumlagerung auf eine funktionelle Gruppe verzichtet werden konnte, da die  -Spaltungen und Wasserstoffumlagerungen
nur für nicht konjugierte Radikale zugelassen wurden. Dies ist ein einfacher Weg, dies für die verwendeten Beispiele zu erreichen, besitzt aber keine allgemeine Gültigkeit. Da diese Regeln ohnehin nur dazu geeignet sind, zu zeigen, daß so
Massenspektren simuliert werden können, aber aus mehreren Gründen (siehe 6.10) nicht breiter eingesetzt werden können, wurde auf eine diesbezügliche Verbesserung verzichtet.
-Spaltungen und Wasserstoffumlagerungen
nur für nicht konjugierte Radikale zugelassen wurden. Dies ist ein einfacher Weg, dies für die verwendeten Beispiele zu erreichen, besitzt aber keine allgemeine Gültigkeit. Da diese Regeln ohnehin nur dazu geeignet sind, zu zeigen, daß so
Massenspektren simuliert werden können, aber aus mehreren Gründen (siehe 6.10) nicht breiter eingesetzt werden können, wurde auf eine diesbezügliche Verbesserung verzichtet.
Wenn diese Ergänzung gewünscht wird, müßten die  -Spaltung und die Wasserstoffumlagerung auch für konjugierte Radikale ergänzt werden und entstehende Radikale nach Möglichkeit konjugiert werden. Für
die Umlagerungszähler würden im Regelteil INIT_RULES entsprechende genealogische Variablen angemeldet, die auch zur Hashcodeberechnung herangezogen werden. Im Teil PREP_ROOT werden sie initialisiert und alle Reaktionstypen außer der Wasserstoffumlagerung
werden sie zurücksetzen. Eine Wasserstoffumlagerung wird nur erlaubt, wenn der Zähler das Maximum von eins noch nicht erreicht hat und dann inkrementiert.
-Spaltung und die Wasserstoffumlagerung auch für konjugierte Radikale ergänzt werden und entstehende Radikale nach Möglichkeit konjugiert werden. Für
die Umlagerungszähler würden im Regelteil INIT_RULES entsprechende genealogische Variablen angemeldet, die auch zur Hashcodeberechnung herangezogen werden. Im Teil PREP_ROOT werden sie initialisiert und alle Reaktionstypen außer der Wasserstoffumlagerung
werden sie zurücksetzen. Eine Wasserstoffumlagerung wird nur erlaubt, wenn der Zähler das Maximum von eins noch nicht erreicht hat und dann inkrementiert.
Die Regeln wurden überdies so geschrieben, daß die Wasserstoffumlagerungen an- und abgeschaltet werden können. Dies geschieht durch den Aufrufparameter -V. Mit -Vhuml#1 übergibt EROS7 eine int-Variable unter dem
Namen huml an die Regeln. Diese testen, ob diese Variable übergeben wurde, welchen Wert sie hat und aktiviert bzw. deaktiviert abhängig davon den Reaktionstyp der Wasserstoffumlagerung. Mit dieser Technik lassen sich Steuerungsmechanismen in die Regeln
einbauen. Hierfür können beliebige Variablen der Typen int und double eingesetzt werden.
#include <iostream.h>
#include rules.h"
#include rh_ilist.h"
#include nn_bpg.h"
#include <math.h>
int get_iprop (rx_eifc *eifc, const char* prop_name, int idx1=0, int idx2=0);
double IP (rx_eifc *eifc, int iels);
int lp_at_pis (rx_eifc *eifc, int iel);
int deconjugate_pis(rx_eifc *eifc, int atom1, int atom2);
RULEOK rules_cc (ostream &print, rx_eifc *eifc, rx_datex *val, int irule, OP op, int sub_op, int test, int trace) {
static double prob=0.;
static int const *level=0;
static int radic_els=0;
static const int *huml=0;
static int def_huml=0;
static nn_bpg nn_alpha, nn_onium, nn_co;
switch(irule) {
case GLOBAL:
switch(op) {
case INIT_RULES: {
val->putco(name","MS-rules for EROS7");
val->putco(date","23.07.97");
static const int input_phase[]={ 0 };
static const int phases_of_reactor[]={ 1, 0 };
static const int phase_property[]={ MONOMOLEC };
static const int phase_contacts[]={ 0 };
static const int input_together[]={ 0 };
val->putco(input_phase_for_reactors", input_phase);
val->putco(phases_per_reactor", phases_of_reactor);
val->putco(phase_property", phase_property);
val->putco(phase_contacts", phase_contacts);
val->putco(use_all_educts_together", input_together);
val->putco(output_phase", 1);
static const char *const kin_mode[]={ prob_kin" };
val->putco(kinetic_model", kin_mode);
val->putco(minimal_concentration", 1.e-5);
val->put(reactivity", &prob);
val->get(level", &level);
val->get(huml", &huml);
if (!(*huml)) huml=&def_huml;
if (!nn_alpha.init(rnnrul3x.dat")) return BAD;
if (!nn_onium.init(rnnrul6zz.dat")) return BAD;
if (!nn_co.init(rnnrul7z.dat")) return BAD;
return OK;}
case PREP_ROOT:
return OK;
case PREP_EDUCT: {
radic_els = 0;
int iel, nel, nelec;
eifc->prop(nel, E_N_ELECSYSS");
for (iel = 1; iel<= nel; iel++) {
eifc->prop(nelec, EL_ELECTRONS",iel);
if (nelec%2) {radic_els = iel; break;}
}
if (*level == 1) return OK;
int charge=get_iprop(eifc, E_CHARGE");
if (charge == 0) return BAD;
return OK;}
case DISTRIBFUNC:
return OK;
case FINISH:
return OK;
default:
return BAD;
}
case 1:
switch(op) {
case RULE_INFO: {
static const char *const attrib[]={ rearrangement", NULL };
static const int center[]={ 0 };
val->putco(rule_name", Ionisation");
val->putco(attributes", attrib);
val->putco(center_connectivity", center);
return OK;}
case CONSTR:
switch(sub_op) {
case 0: {
if (get_iprop(eifc, E_CHARGE")) return BAD;
return OK;}
default:
return BAD;
}
case FUNC: {
int iel, nel, nelec, igr, sgr;
rh_ilist<int> hgroup_idxs;
vector<int> hgroup;
eifc->prop(nel, E_N_ELECSYSS");
for (iel = 1; iel <= nel; iel++) {
eifc->set_active_ens(1);
if (!get_iprop(eifc, EL_IS_SIGMA", iel)) {
eifc->prop(hgroup, EL_HGROUP", iel);
igr = hgroup[0];
sgr = hgroup[1];
eifc->prop(nelec,"EL_ELECTRONS", iel);
if ((!hgroup_idxs.at_idx(igr)) && (nelec)) {
if (!lp_at_pis(eifc, iel)) {
prob = 1e11 * exp(-2.*IP(eifc, iel));
eifc->change_electrons(iel, -1);
eifc->symmetry_factor = sgr; // bzw. = sgr
print << Ioni: prob = << prob << endl;
eifc->save_active_ens_as_product();
hgroup_idxs.append(igr);
}
}
}
}
return OK;}
default:
return BAD;
}
case 2:
switch(op) {
case RULE_INFO: {
static const char *const attrib[]={ NULL };
static const int center[]={ 1, 2, 0, 2, 3, 0, 0 };
val->putco(rule_name", Alpha cleavage");
val->putco(attributes", attrib);
val->putco(center_connectivity", center);
return OK;}
case CONSTR:
switch(sub_op) {
case 0: {
if (!get_iprop(eifc, E_RADICAL")) return BAD;
vector<int> atoms;
if (!radic_els) return BAD;
eifc->prop(atoms, EL_ATOMS", radic_els);
if (atoms.size() > 1) return BAD;
return OK;}
case 1:
if (!get_iprop(eifc, A_RADICCENTER", eifc->center(1))) return BAD;
return OK;
case 2: {
vector<int> els;
eifc->prop(els, N_ELECSYSS", eifc->center(1), eifc->center(2));
if (els.size() > 2) return BAD;
if ((els[0] == radic_els) || (els[els.size()] == radic_els)) return BAD;
return OK;}
case 3: {
//if (get_iprop(eifc, A_ELEMENT", eifc->center(3)) == 1) return BAD;
vector<int> els;
eifc->prop(els, N_ELECSYSS", eifc->center(2), eifc->center(3));
if (els.size() > 2) return BAD;
return OK;}
default:
return BAD;
}
case FUNC: {
int issys, c1, c2, c3, iel1, iel2, imol, elem;
double chrg;
c1 = eifc->center(1);
c2 = eifc->center(2);
c3 = eifc->center(3);
nn_alpha.set_vals(0, eifc);
issys = deconjugate_pis(eifc, c2, c3);
if (!issys) return BAD;
eifc->split_elsy(issys, c2, c3, iel1, iel2, 1);
eifc->combine_elsys(radic_els, iel1, es_pi);
eifc->prop(imol, A_MOLECULE", c1);
if (!get_iprop(eifc, M_CHARGE", imol)) return BAD;
nn_alpha.set_vals(1, eifc);
prob = nn_alpha.get_val();
eifc->prop(elem, A_ELEMENT", c3);
eifc->prop(chrg, A_CHARGECENTER", c1);
if ((elem == 1) && (chrg < 0.1)) prob *= 0.06;
print << alpha: prob = << prob << endl;
if (prob < 0.05) return BAD;
return OK;}
default:
return BAD;
}
case 3:
switch(op) {
case RULE_INFO: {
static const char *const attrib[]={ NULL };
static const int center[]={ 1, 2, 0, 3, 4, 0, 0 };
val->putco(rule_name", Onium reaction");
val->putco(attributes", attrib);
val->putco(center_connectivity", center);
return OK;}
case CONSTR:
switch(sub_op) {
case 0:
if (get_iprop(eifc, E_RADICAL")) return BAD;
return OK;
case 1: {
int c1, elem, i, n, nel, nat, iel;
double chrg;
vector<int> els, atoms;
c1 = eifc->center(1);
eifc->prop(elem, A_ELEMENT", c1);
if ((elem == 1) || (elem == 6)) return BAD;
eifc->prop(chrg, A_CHARGECENTER", c1);
if (chrg <= 0.) return BAD;
eifc->prop(els, A_ELECSYSS", c1);
n = els.size();
for (i=0; i<n; i++) {
iel = els[i];
if (!get_iprop(eifc, EL_IS_SIGMA", iel)) {
eifc->prop(atoms, EL_ATOMS", iel);
nat = atoms.size();
eifc->prop(nel, EL_ELECTRONS", iel);
if ((nat > 1) && (nel > 1)) return OK;
}
}
return BAD;}
case 2: {
int c1=eifc->center(1), c2=eifc->center(2);
if (get_iprop(eifc, A_ELEMENT", c2) != 6) return BAD;
vector<int> els;
eifc->prop(els, N_ELECSYSS", c1, c2);
if (els.size() != 1) return BAD;
return OK;}
case 3: {
int c1=eifc->center(1), c2=eifc->center(2), c3=eifc->center(3), idist, idist2;
if (get_iprop(eifc, A_ELEMENT", c3) != 6) return BAD;
eifc->prop(idist, N_DIST_INDEX", c2, c3);
if (idist > 4) return BAD;
eifc->prop(idist2, N_DIST_INDEX", c1, c3);
if (idist2 < idist) return BAD;
return OK;}
case 4:
if (get_iprop(eifc, A_ELEMENT", eifc->center(4)) != 1) return BAD;
return OK;
default:
return BAD;
}
case FUNC: {
int c1, c2, c3, c4;
c1 = eifc->center(1);
c2 = eifc->center(2);
c3 = eifc->center(3);
c4 = eifc->center(4);
nn_onium.set_vals(0, eifc);
eifc->change_bond_order(c1, c2, -1);
eifc->change_bond_order(c3, c4, -1);
eifc->change_bond_order(c1, c4, 1);
eifc->change_bond_order(c2, c3, 1);
nn_onium.set_vals(1, eifc);
prob = 0.5 * nn_onium.get_val();
print << onium: prob = << prob << endl;
if (prob < 0.05) return BAD;
return OK;}
default:
return BAD;
}
case 4:
switch(op) {
case RULE_INFO: {
static const char *const attrib[]={ NULL };
static const int center[]={ 1, 2, 0, 2, 3, 0, 0 };
val->putco(rule_name", Carbonyl elimination");
val->putco(attributes", attrib);
val->putco(center_connectivity", center);
return OK;}
case CONSTR:
switch(sub_op) {
case 0:
if (radic_els) return BAD;
return OK;
case 1: {
double chrg;
int elem = get_iprop(eifc, A_ELEMENT", eifc->center(1));
if ((elem != 8) && (elem != 7)) return BAD;
eifc->prop(chrg, A_CHARGECENTER", eifc->center(1));
if (chrg <= 0.) return BAD;
return OK;}
case 2: {
int c1 = eifc->center(1), c2 = eifc->center(2);
if (get_iprop(eifc, A_ELEMENT", c2) != 6) return BAD;
vector<int> els;
eifc->prop(els, N_ELECSYSS", c1, c2);
if (els.size() != 3) return BAD;
return OK;}
case 3:
if (get_iprop(eifc, A_ELEMENT", eifc->center(3)) == 1) return BAD;
return OK;
default:
return BAD;
}
case FUNC: {
int issys, c2, c3, iel1, iel2;
c2 = eifc->center(2);
c3 = eifc->center(3);
nn_co.set_vals(0, eifc);
issys = deconjugate_pis(eifc, c2, c3);
if (!issys) return BAD;
eifc->split_elsy(issys, c2, c3, iel1, iel2, 2);
nn_co.set_vals(1, eifc);
prob = 0.5 * nn_co.get_val();
print << co: prob = << prob << endl;
if (prob < 0.05) return BAD;
return OK;}
default:
return BAD;
}
case 5:
switch(op) {
case RULE_INFO: {
static const char *const attrib[]={ NULL };
static const int center[]={ 1,2,0, 2,3,0, 3,4,0, 4,5,0, 5,6,0, 0 };
val->putco(rule_name", McLafferty reaction");
val->putco(attributes", attrib);
val->putco(center_connectivity", center);
return OK;}
case CONSTR:
switch(sub_op) {
case 0: {
if (!radic_els) return BAD;
vector<int> atoms;
eifc->prop(atoms, EL_ATOMS", radic_els);
if (atoms.size() != 1) return BAD;
return OK;}
case 1: {
int c1 = eifc->center(1), elem, i, n;
eifc->prop(elem, A_ELEMENT", c1);
if (elem != 8) return BAD;
vector<int> els;
eifc->prop(els, A_ELECSYSS", c1);
n = els.size();
for (i=0; i<n; i++) {
if (els[i] == radic_els) return OK;
}
return BAD;}
case 2: {
vector<int> els;
eifc->prop(els, N_ELECSYSS", eifc->center(1), eifc->center(2));
if (els.size() != 2) return BAD;
return OK;}
case 3:
if (get_iprop(eifc, A_ELEMENT", eifc->center(3)) != 6) return BAD;
return OK;
case 4:
return OK;
case 5:
return OK;
case 6:
if (get_iprop(eifc, A_ELEMENT", eifc->center(6)) != 1) return BAD;
return OK;
default:
return BAD;
}
case FUNC: {
int i, n, c1, c2, c3, c4, c5, c6, ipis, iel1, iel2, iel3, iel4, nhn, issys;
vector<int> v;
c1 = eifc->center(1);
c2 = eifc->center(2);
c3 = eifc->center(3);
c4 = eifc->center(4);
c5 = eifc->center(5);
c6 = eifc->center(6);
nhn = 0;
eifc->prop(v, A_NEIGHBORS", c5);
n = v.size();
for (i=0; i<n; i++) {
if (get_iprop(eifc, A_ELEMENT", v[i]) != 1) nhn++;
}
issys = deconjugate_pis(eifc, c3, c4);
if (!issys) return BAD;
eifc->prop(v, N_ELECSYSS", c1, c2);
n = v.size();
for (i=0; i<n; i++) {
if (!get_iprop(eifc, EL_IS_SIGMA", v[i])) ipis = v[i];
}
eifc->prop(v, N_ELECSYSS", c5, c6);
eifc->split_elsy(v[0], c5, c6, iel1, iel2, 1);
eifc->split_elsy(issys, c3, c4, iel3, iel4, 1);
eifc->combine_elsys(radic_els, iel2, es_sigma);
eifc->combine_elsys(iel1, iel4, es_pi);
eifc->combine_elsys(ipis, iel3, es_pi);
prob = 0.8361;
if (nhn == 1) prob = 0.23;
print << ml: prob = << prob << endl;
if (prob < 0.05) return BAD;
return OK;}
default:
return BAD;
}
case 6:
switch(op) {
case RULE_INFO: {
static const char *const attrib[]={ rearrangement", NULL };
static const int center[]={ 2, 3, 0, 0 };
val->putco(rule_name", Hydrogen rearrangement");
val->putco(attributes", attrib);
val->putco(center_connectivity", center);
return OK;}
case CONSTR:
switch(sub_op) {
case 0: {
if (!(*huml)) return BAD;
if (!radic_els) return BAD;
vector<int> atoms;
eifc->prop(atoms, EL_ATOMS", radic_els);
if (atoms.size() != 1) return BAD;
return OK;}
case 1: {
int c1 = eifc->center(1), i, n;
vector<int> els;
eifc->prop(els, A_ELECSYSS", c1);
n = els.size();
for (i=0; i<n; i++) {
if (els[i] == radic_els) return OK;
}
return BAD;}
case 2: {
int idist, c1 = eifc->center(1), c2 = eifc->center(2);
eifc->prop(idist, N_DIST_INDEX", c1, c2);
if (get_iprop(eifc, A_ELEMENT", c1) == 6) {
if ((idist < 3) || (idist > 5)) return BAD;
} else {
if (idist != 2) return BAD;
}
return OK;}
case 3:
if (get_iprop(eifc, A_ELEMENT", eifc->center(3)) != 1) return BAD;
return OK;
default:
return BAD;
}
case FUNC: {
int iel1, iel2, c1, c2, c3, elem, idist;
vector<int> els;
c1 = eifc->center(1);
c2 = eifc->center(2);
c3 = eifc->center(3);
elem = get_iprop(eifc, A_ELEMENT", c1);
eifc->prop(idist, N_DIST_INDEX", c1, c2);
eifc->prop(els, N_ELECSYSS", c2, c3);
eifc->split_elsy(els[0], c2, c3, iel1, iel2, 1);
eifc->combine_elsys(radic_els, iel2, es_sigma);
if (elem == 6) {
switch(idist) {
case 3:
prob = 0.1;
break;
case 4:
prob = 0.3;
break;
case 5:
prob = 0.05;
break;
default:
return BAD;
}
} else {
prob = 0.5;
}
eifc->symmetry_factor = 1; // oder nicht?
print << h-uml: prob = << prob << endl;
return OK;}
default:
return BAD;
}
default:
return BAD;
}
}
// --------------------------------------------------------------------------
int get_iprop(rx_eifc *eifc, const char* prop_name, int idx1, int idx2) {
int ivar=0;
eifc->prop(ivar, prop_name, idx1, idx2);
return ivar;
}
double IP(rx_eifc *eifc, int iels) {
return 10.;
}
int lp_at_pis(rx_eifc *eifc, int iel) {
int iel2, iat, nels, nat, i, j, nelec;
if (get_iprop(eifc, EL_IS_SIGMA", iel)) return 0;
vector<int> atoms, elsyss, atoms2;
eifc->prop(atoms,"EL_ATOMS", iel);
nat = atoms.size();
if (nat == 1) return 0;
for (i=0; i<nat; i++) {
iat = atoms[i];
eifc->prop(elsyss, A_ELECSYSS", iat);
nels = elsyss.size();
for (j=0; j<nels; j++) {
iel2 = elsyss[j];
if (!get_iprop(eifc, EL_IS_SIGMA", iel2)) {
eifc->prop(atoms2, EL_ATOMS", iel2);
eifc->prop(nelec, EL_ELECTRONS", iel2);
if ((nelec)&&(atoms2.size()==1)) return 1;
}
}
}
return 0;
}
int deconjugate_pis(rx_eifc *eifc, int atom1, int atom2) {
int issys=0, i, n, iel1, iel2;
vector<int> elsys, etyp;
eifc->prop(elsys, N_ELECSYSS", atom1, atom2);
n = elsys.size();
etyp.resize(n);
for (i=0; i<n; i++) {
etyp[i] = get_iprop(eifc, EL_IS_SIGMA", elsys[i]);
}
for (i=0; i<n; i++) {
if (etyp[i]) {
issys = elsys[i];
} else {
if (eifc->split_elsy(elsys[i], atom1, atom2, iel1, iel2, -1)) return 0;
}
}
return issys;
}
Der Inhalt der folgenden drei Dateien sind die Gewichte, Skalierungen und Parameter für die Backpropagationnetze. Zum möglichst leichten Umstieg von EROS6 auf EROS7 wurde am Dateiformat für die Backpropagationnetze nur die Angabe der Namen der physikochemischen Parameter geändert. /NN_PARM ist die Angabe der Parameter in VERGIL und wird von EROS7 nicht gelesen und ist nur aus Kompatibilitätsgründen enthalten. Die restlichen Abschnitte der Datei für die Backpropagationnetze sind identisch. Für EROS7 sind die unter /NN_PARM7 angegebenen Eigenschaften maßgebend. Die vier Zahlen dahinter sind die zwei Indizes, die auf null gesetzt sind, wenn sie nicht benötigt werden, eine Zahl für das Ensemble, von dem die Eigenschaft genommen werden soll (0 = Edukt, 1 = Produkt bzw. erstes Zwischenprodukt usw.). Die letzte gibt den Datentyp an (0 = ganzzahlig / int, 1 = Fließkommazahl / double). /NN_ARCHT, /NN_CONNS und /NN_SCAL haben das gleiche Format wie die Netze von EROS6. /NN_ARCHT folgt eine Zeile mit der Zahl der versteckten Schichten (hidden layers) und in der nächsten Zeile die Zahl der Neuronen für jede Schicht ohne Bias beginnend mit der Eingabeschicht. /NN_SCAL gibt nach der fortlaufenden Numerierung die minimalen und maximalen Werte der Eigenschaft während des Trainings an. Die Eigenschaften zwischen diesen Werten werden von den Eingabeneuronen in einen Wertebereich von null bis eins skaliert. Diese Skalierung ist für alle Inputneuronen angegeben und wird von der Skalierung der Ausgabeneuronen gefolgt. Hier ist der Bereich angegeben, in den die Werte zwischen null und eins skaliert werden. Unter /NN_CONNS sind sämtliche Gewichte des Netzes angegeben und zwar schichtenweise, beginnend mit den Gewichten zwischen der Eingabeschicht und der ersten versteckten Schicht. Die Gewichte kommen dann für ein Netz mit einem hidden layer und einem Ausgabeneuron in der Reihenfolge: input1 - hidden1, input2 - hidden1, ..., bias - hidden1, input1 - hidden2, ..., bias - hidden_last, hidden1 - output, ..., bias - output.
 -Spaltung:
-Spaltung:
/NN_ARCHT 2 2
1
12 3 1
/NN_PARM 16 16
PARAMETER EDUKT:
1 ATOM 1 ENSIG *
2 ATOM 1 FREEEL *
3 ATOM 1 NEIGHBOR *
4 BOND 2 3 TBDE *
5 ENSEMBLE EDELTAHF *
STOP
PARAMETER PRODUKT:
6 ATOM 2 APOLARIZABILITY *
7 ATOM 2 ENPI *
8 ATOM 2 QSIG *
9 ATOM 3 APOLARIZABILITY *
10 ATOM 3 QSIG *
11 BOND 2 1 PSTAB *
12 ENSEMBLE EDELTAHF *
END
/NN_PARM7 12 12
A_ENSIG 1 0 0 1
A_FREEEL 1 0 0 0
A_NEIGHBOR 1 0 0 0
B_TBDE 2 3 0 1
E_DELTAHF 0 0 0 1
A_POLARIZ 2 0 1 1
A_ENPI 2 0 1 1
A_QSIG 2 0 1 1
A_POLARIZ 3 0 1 1
A_QSIG 3 0 1 1
B_PSTAB 2 1 1 1
E_DELTAHF 0 0 1 1
/NN_SCAL 13 13
1 8.968300 15.304200
2 1.000000 5.000000
3 1.000000 3.000000
4 -92.822502 480.858002
5 257.174805 945.775330
6 2.125000 9.201000
7 6.641700 10.931600
8 -0.055000 0.552900
9 0.386000 6.782400
10 -0.252800 0.000000
11 0.000000 7.455200
12 -15.910600 1213.183960
13 0.000000 0.995000
/NN_CONNS 43 43
1.139383
1.473840
1.395796
-1.247808
-0.440537
-1.299575
-1.419531
3.151312
-2.142884
0.546493
-6.229643
-3.132250
-0.539434
-0.682565
-4.904122
2.436973
-0.518647
5.592857
4.013568
3.796338
2.633508
4.288496
-2.034354
0.601260
-3.548747
-6.780631
-0.523646
6.964950
-3.849618
-0.455457
-0.303718
2.028105
-3.101313
1.117309
4.066958
0.729223
-2.652695
-2.812138
-0.675825
6.165740
-6.543190
-10.318967
5.170370
/END 0 0
/NN_ARCHT 2 2
1
11 2 1
/NN_PARM 15 15
PARAMETER EDUKT:
1 ATOM 1 ENPI *
2 ATOM 1 FREEEL *
3 ATOM 1 NONHNEIGHBOR *
4 ATOM 1 QPI *
5 ATOM 2 NONHNEIGHBOR *
6 BOND 1 2 DENSIG *
7 BOND 1 2 DQSIG *
8 ENSEMBLE EDELTAHF *
STOP
PARAMETER PRODUKT:
9 ATOM 1 APOLARIZABILITY *
10 ATOM 1 LPSTAB *
11 ATOM 4 QTOT *
END
/NN_PARM7 11 11
A_ENPI 1 0 0 1
A_FREEEL 1 0 0 0
A_NONHNEIGHBOR 1 0 0 0
A_QPI 1 0 0 1
A_NONHNEIGHBOR 2 0 0 0
B_DENSIG 1 2 0 1
B_DQSIG 1 2 0 1
E_DELTAHF 0 0 0 1
A_POLARIZ 1 0 1 1
A_LPSTAB 1 0 1 1
A_QTOT 4 0 1 1
/NN_SCAL 12 12
1 9.455500 11.616000
2 0.000000 2.000000
3 2.000000 3.000000
4 0.100800 0.242300
5 2.000000 4.000000
6 0.088200 2.725800
7 -0.474800 0.011600
8 -98.436996 656.533691
9 2.559000 7.090800
10 0.000000 10.057100
11 -0.119200 -0.091800
12 0.000000 0.999000
/NN_CONNS 27 27
-0.631653
-2.841740
3.425206
-1.077220
-0.205757
3.651624
-3.584734
0.577689
-2.491549
-0.240271
1.744900
0.944103
-0.749909
2.701843
5.886912
-2.439176
3.877181
-3.218500
-9.040503
-5.702037
-0.892885
6.837634
2.773469
-2.080549
-9.147391
5.189584
10.084118
/END 0 0
/NN_ARCHT 2 2
1
2 1 1
/NN_PARM 6 6
PARAMETER EDUKT:
1 ATOM 2 ENPI *
2 BOND 3 2 PSTAB *
STOP
PARAMETER PRODUKT:
END
/NN_PARM7 2 2
A_ENPI 2 0 0 1
B_PSTAB 3 2 0 1
/NN_SCAL 3 3
1 9.305900 10.108800
2 0.000000 7.876400
3 0.048000 1.000000
/NN_CONNS 5 5
-3.467265
2.447763
4.537941
-3.087231
6.941255
/END 0 0
![[Prev]](p_prev.gif)
![[Next]](p_next.gif)